RDC Integration
BiodivPortal is part of the RDC mediation layer. It stores and manages a set of terminologies and standard schemas useful for transforming data and metadata into a standard format for processing and storage in the RDC Semantic Layer. BiodivPortal offers among others, services to (i) annotate texts with terms (ii) establish links between semantically related terms (iii) make terms referenceable via stable URIs and (iv) make term versioning transparent.
Semantic search capabilities will be enabled through the Semantic Lookup Index, which stores information about concept synonyms, acronyms, common names as well as broader terms.
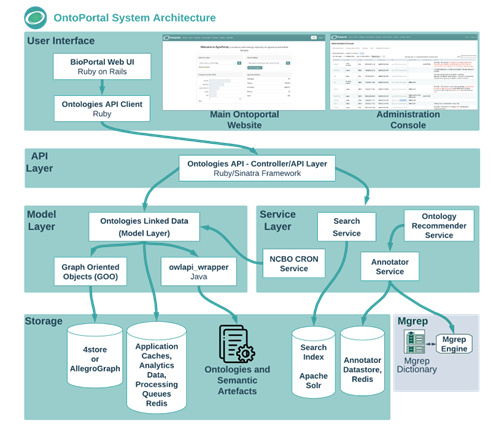
As members of the OntoPortal Alliance, we are maintaining and further developing our own instance of the OntoPortal technology dedicated to biodiversity related terminologies, called BiodivPortal. In our context, a terminology refers to any terminological resource, this can be a formal ontology like ENVO, a taxonomy like ITIS, or any useful collections of terms (e.g. locations available via a geographical database like Geonames). OntoPortal is a generic technology to build terminology repositories, catalogues and services. The system architecture of OntoPortal is structured in several layers:
- The storage layer contains a triplestore which saves each terminology in a distinct graph, as well as other data (metadata records, mappings, users, etc.). This layer also includes: (i) a storage for application caches and the Annotator dictionary datastore; (ii) a Solr search engine to index terminology content for retrieval with the Search service.
- The model layer implements all the mechanisms to parse the terminology source files using the OWL API and retrieve them from the triple-store using a built-in Object-Relational-Mapping library (GOO).
- The service layer implements the core OntoPortal services: Search, Annotator and Recommender.
- The API layer implements a unified API for all the models (e.g., Class, Instance, Ontology, Submission, Mapping, Review, Note, User) and services supported by OntoPortal. The API returns as default a JSON-LD format.
- The user interface offers a set of various views to display and use the services and components built in the API layer. The UI is customized for logged-in users and for groups/organizations that display their own subset of resources. Administrators of the OntoPortal instance have access to an additional administration console to monitor, and manage the content of the portal.
Getting started
BiodivPortal is available on http://biodivportal.gfbio.org. Users can register to get an account and an API key. As described in the following sections, users can explore, contibute or consume terminologies through the User Interface (UI), Developers can programatically access all available terminologies and services.
User Guide
BiodivPortal provides access through its User Interface to useful biodiversity related terminologies (i.e. vocabularies, taxonomies and ontologies) as well as tools for working with them. It allows users to:
- Browse the repository and terminologies content
- Search for a term across multiple terminologies
- Browse mappings between terms in different terminologies
- Receive recommendations on which terminologies are most relevant for a corpus
- Annotate text with terms from terminologies
Logged-in users can additionally:
- Submit their own terminology in a private or public mode
- Add mappings between terms
- Add comments or change suggestions for a terminology or a specific term
A comprehensive user documentation can be found on the BioPortal wiki page: https://www.bioontology.org/wiki/BioPortal_Help
Developer Guide
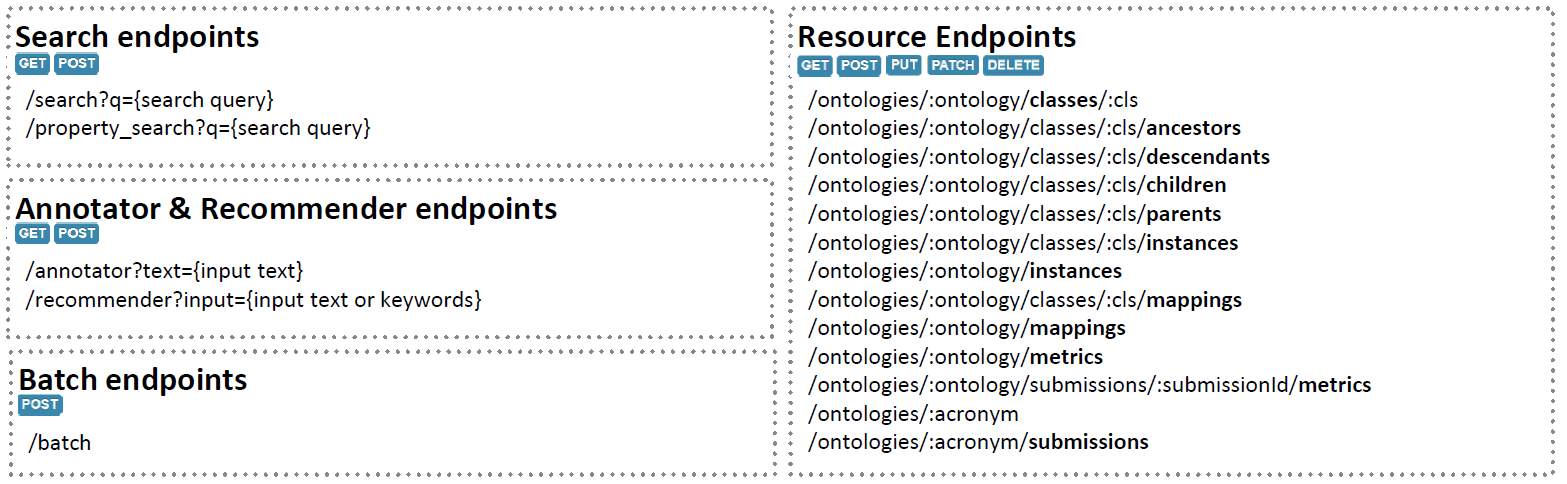
All information available through the User Interface is also available through the Web Service REST API: https://data.biodivportal.gfbio.org/. In the following is an overview of the main available API endpoints:
The complete BiodivPortal API documentation can be found here: https://data.biodivportal.gfbio.dev/documentation
References
Links:
- Main instance: http://biodivportal.gfbio.org
- BiodivPortal Code: https://github.com/BiodivPortal
- RDB2RDF tool: https://github.com/biodivportal/rdb2rdf